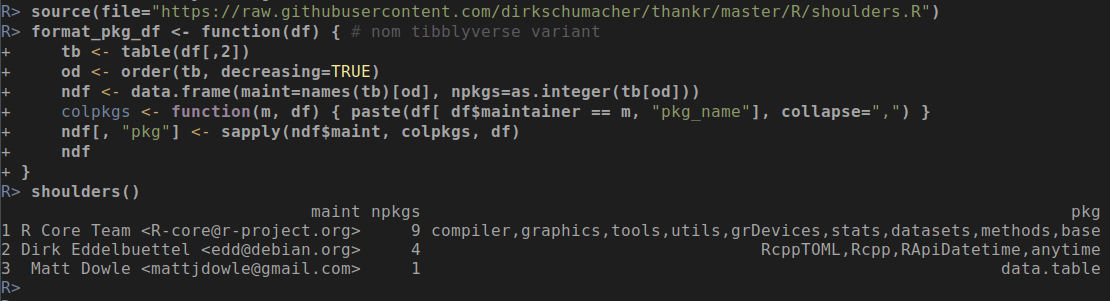
Now, to actually use R-devel you obviously need to have it accessible. There are a myriad of ways to achieve that: just compile it locally as I have done for years, use a Docker image as I showed in the post -- or be creative with eg Travis or win-builder both of which give you access to R-devel if you're clever about it.
But as no good deed goes unpunished, I was of course told off today for showing a Docker example as Docker was not "Easy". I think the formal answer to that is baloney. But we leave that aside, and promise to discuss setting up Docker at another time.
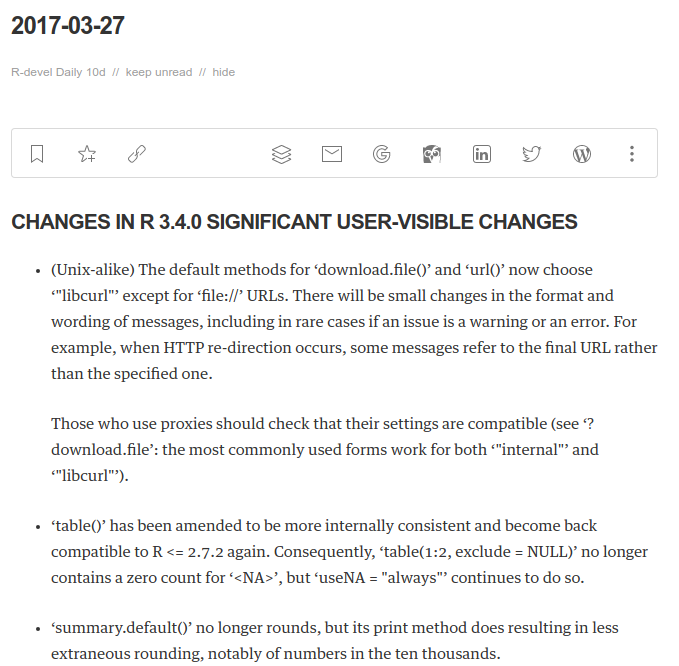
R is after all ... just R. So below please find a script you can save as, say, ~/bin/pnrrs.r. And calling it---even with R-release---will generate the same code snippet as I showed via Docker. Call it a one-off backport of the new helper function -- with a half-life of a few weeks at best as we will have R 3.4.0 as default in just a few weeks. The script will then reduce to just the final line as the code will be present with R 3.4.0.
#!/usr/bin/r
library(tools)
.find_calls_in_package_code <- tools:::.find_calls_in_package_code
.read_description <- tools:::.read_description
## all what follows is from R-devel aka R 3.4.0 to be
package_ff_call_db <- function(dir) {
## A few packages such as CDM use base::.Call
ff_call_names <- c(".C", ".Call", ".Fortran", ".External",
"base::.C", "base::.Call",
"base::.Fortran", "base::.External")
predicate <- function(e) {
(length(e) > 1L) &&
!is.na(match(deparse(e[[1L]]), ff_call_names))
}
calls <- .find_calls_in_package_code(dir,
predicate = predicate,
recursive = TRUE)
calls <- unlist(Filter(length, calls))
if(!length(calls)) return(NULL)
attr(calls, "dir") <- dir
calls
}
native_routine_registration_db_from_ff_call_db <- function(calls, dir = NULL, character_only = TRUE) {
if(!length(calls)) return(NULL)
ff_call_names <- c(".C", ".Call", ".Fortran", ".External")
ff_call_args <- lapply(ff_call_names,
function(e) args(get(e, baseenv())))
names(ff_call_args) <- ff_call_names
ff_call_args_names <-
lapply(lapply(ff_call_args,
function(e) names(formals(e))), setdiff,
"...")
if(is.null(dir))
dir <- attr(calls, "dir")
package <- # drop name
as.vector(.read_description(file.path(dir, "DESCRIPTION"))["Package"])
symbols <- character()
nrdb <-
lapply(calls,
function(e) {
if (startsWith(deparse(e[[1L]]), "base::"))
e[[1L]] <- e[[1L]][3L]
## First figure out whether ff calls had '...'.
pos <- which(unlist(Map(identical,
lapply(e, as.character),
"...")))
## Then match the call with '...' dropped.
## Note that only .NAME could be given by name or
## positionally (the other ff interface named
## arguments come after '...').
if(length(pos)) e <- e[-pos]
## drop calls with only ...
if(length(e) < 2L) return(NULL)
cname <- as.character(e[[1L]])
## The help says
##
## '.NAME' is always matched to the first argument
## supplied (which should not be named).
##
## But some people do (Geneland ...).
nm <- names(e); nm[2L] <- ""; names(e) <- nm
e <- match.call(ff_call_args[[cname]], e)
## Only keep ff calls where .NAME is character
## or (optionally) a name.
s <- e[[".NAME"]]
if(is.name(s)) {
s <- deparse(s)[1L]
if(character_only) {
symbols <<- c(symbols, s)
return(NULL)
}
} else if(is.character(s)) {
s <- s[1L]
} else { ## expressions
symbols <<- c(symbols, deparse(s))
return(NULL)
}
## Drop the ones where PACKAGE gives a different
## package. Ignore those which are not char strings.
if(!is.null(p <- e[["PACKAGE"]]) &&
is.character(p) && !identical(p, package))
return(NULL)
n <- if(length(pos)) {
## Cannot determine the number of args: use
## -1 which might be ok for .External().
-1L
} else {
sum(is.na(match(names(e),
ff_call_args_names[[cname]]))) - 1L
}
## Could perhaps also record whether 's' was a symbol
## or a character string ...
cbind(cname, s, n)
})
nrdb <- do.call(rbind, nrdb)
nrdb <- as.data.frame(unique(nrdb), stringsAsFactors = FALSE)
if(NROW(nrdb) == 0L || length(nrdb) != 3L)
stop("no native symbols were extracted")
nrdb[, 3L] <- as.numeric(nrdb[, 3L])
nrdb <- nrdb[order(nrdb[, 1L], nrdb[, 2L], nrdb[, 3L]), ]
nms <- nrdb[, "s"]
dups <- unique(nms[duplicated(nms)])
## Now get the namespace info for the package.
info <- parseNamespaceFile(basename(dir), dirname(dir))
## Could have ff calls with symbols imported from other packages:
## try dropping these eventually.
imports <- info$imports
imports <- imports[lengths(imports) == 2L]
imports <- unlist(lapply(imports, `[[`, 2L))
info <- info$nativeRoutines[[package]]
## Adjust native routine names for explicit remapping or
## namespace .fixes.
if(length(symnames <- info$symbolNames)) {
ind <- match(nrdb[, 2L], names(symnames), nomatch = 0L)
nrdb[ind > 0L, 2L] <- symnames[ind]
} else if(!character_only &&
any((fixes <- info$registrationFixes) != "")) {
## There are packages which have not used the fixes, e.g. utf8latex
## fixes[1L] is a prefix, fixes[2L] is an undocumented suffix
nrdb[, 2L] <- sub(paste0("^", fixes[1L]), "", nrdb[, 2L])
if(nzchar(fixes[2L]))
nrdb[, 2L] <- sub(paste0(fixes[2L]), "$", "", nrdb[, 2L])
}
## See above.
if(any(ind <- !is.na(match(nrdb[, 2L], imports))))
nrdb <- nrdb[!ind, , drop = FALSE]
## Fortran entry points are mapped to l/case
dotF <- nrdb$cname == ".Fortran"
nrdb[dotF, "s"] <- tolower(nrdb[dotF, "s"])
attr(nrdb, "package") <- package
attr(nrdb, "duplicates") <- dups
attr(nrdb, "symbols") <- unique(symbols)
nrdb
}
format_native_routine_registration_db_for_skeleton <- function(nrdb, align = TRUE, include_declarations = FALSE) {
if(!length(nrdb))
return(character())
fmt1 <- function(x, n) {
c(if(align) {
paste(format(sprintf(" {\"%s\",", x[, 1L])),
format(sprintf(if(n == "Fortran")
"(DL_FUNC) &F77_NAME(%s),"
else
"(DL_FUNC) &%s,",
x[, 1L])),
format(sprintf("%d},", x[, 2L]),
justify = "right"))
} else {
sprintf(if(n == "Fortran")
" {\"%s\", (DL_FUNC) &F77_NAME(%s), %d},"
else
" {\"%s\", (DL_FUNC) &%s, %d},",
x[, 1L],
x[, 1L],
x[, 2L])
},
" {NULL, NULL, 0}")
}
package <- attr(nrdb, "package")
dups <- attr(nrdb, "duplicates")
symbols <- attr(nrdb, "symbols")
nrdb <- split(nrdb[, -1L, drop = FALSE],
factor(nrdb[, 1L],
levels =
c(".C", ".Call", ".Fortran", ".External")))
has <- vapply(nrdb, NROW, 0L) > 0L
nms <- names(nrdb)
entries <- substring(nms, 2L)
blocks <- Map(function(x, n) {
c(sprintf("static const R_%sMethodDef %sEntries[] = {",
n, n),
fmt1(x, n),
"};",
"")
},
nrdb[has],
entries[has])
decls <- c(
"/* FIXME: ",
" Add declarations for the native routines registered below.",
"*/")
if(include_declarations) {
decls <- c(
"/* FIXME: ",
" Check these declarations against the C/Fortran source code.",
"*/",
if(NROW(y <- nrdb$.C)) {
args <- sapply(y$n, function(n) if(n >= 0)
paste(rep("void *", n), collapse=", ")
else "/* FIXME */")
c("", "/* .C calls */",
paste0("extern void ", y$s, "(", args, ");"))
},
if(NROW(y <- nrdb$.Call)) {
args <- sapply(y$n, function(n) if(n >= 0)
paste(rep("SEXP", n), collapse=", ")
else "/* FIXME */")
c("", "/* .Call calls */",
paste0("extern SEXP ", y$s, "(", args, ");"))
},
if(NROW(y <- nrdb$.Fortran)) {
args <- sapply(y$n, function(n) if(n >= 0)
paste(rep("void *", n), collapse=", ")
else "/* FIXME */")
c("", "/* .Fortran calls */",
paste0("extern void F77_NAME(", y$s, ")(", args, ");"))
},
if(NROW(y <- nrdb$.External))
c("", "/* .External calls */",
paste0("extern SEXP ", y$s, "(SEXP);"))
)
}
headers <- if(NROW(nrdb$.Call) || NROW(nrdb$.External))
c("#include <R.h>", "#include <Rinternals.h>")
else if(NROW(nrdb$.Fortran)) "#include <R_ext/RS.h>"
else character()
c(headers,
"#include <stdlib.h> // for NULL",
"#include <R_ext/Rdynload.h>",
"",
if(length(symbols)) {
c("/*",
" The following symbols/expresssions for .NAME have been omitted",
"", strwrap(symbols, indent = 4, exdent = 4), "",
" Most likely possible values need to be added below.",
"*/", "")
},
if(length(dups)) {
c("/*",
" The following name(s) appear with different usages",
" e.g., with different numbers of arguments:",
"", strwrap(dups, indent = 4, exdent = 4), "",
" This needs to be resolved in the tables and any declarations.",
"*/", "")
},
decls,
"",
unlist(blocks, use.names = FALSE),
## We cannot use names with '.' in: WRE mentions replacing with "_"
sprintf("void R_init_%s(DllInfo *dll)",
gsub(".", "_", package, fixed = TRUE)),
"{",
sprintf(" R_registerRoutines(dll, %s);",
paste0(ifelse(has,
paste0(entries, "Entries"),
"NULL"),
collapse = ", ")),
" R_useDynamicSymbols(dll, FALSE);",
"}")
}
package_native_routine_registration_db <- function(dir, character_only = TRUE) {
calls <- package_ff_call_db(dir)
native_routine_registration_db_from_ff_call_db(calls, dir, character_only)
}
package_native_routine_registration_db <- function(dir, character_only = TRUE) {
calls <- package_ff_call_db(dir)
native_routine_registration_db_from_ff_call_db(calls, dir, character_only)
}
package_native_routine_registration_skeleton <- function(dir, con = stdout(), align = TRUE,
character_only = TRUE, include_declarations = TRUE) {
nrdb <- package_native_routine_registration_db(dir, character_only)
writeLines(format_native_routine_registration_db_for_skeleton(nrdb,
align, include_declarations),
con)
}
package_native_routine_registration_skeleton(".") ## when R 3.4.0 is out you only need this line