#50: Introducing ‘almm: Activate-Linux (based) Market Monitor’
Welcome to post 50 in the R4 series.
Today we reconnect to a previous post, namely #36 on pub/sub for live market monitoring with R and Redis. It introduced both Redis as well as the (then fairly recent) extensions to RcppRedis to support the publish-subscibe (“pub/sub”) model of Redis. In short, it manages both subscribing clients as well as producer for live, fast and lightweight data transmission. Using pub/sub is generally more efficient than the (conceptually simpler) ‘poll-sleep’ loops as polling creates cpu and network load. Subscriptions are lighterweight as they get notified, they are also a little (but not much!) more involved as they require a callback function.
We should mention that Redis has a recent fork in Valkey that arose when the former did one of these non-uncommon-among-db-companies licenuse suicides—which, happy to say, they reversed more recently—so that we now have both the original as well as this leading fork (among others). Both work, the latter is now included in several Linux distros, and the C library hiredis used to connect to either is still licensed permissibly as well.
All this came about because Yahoo! Finance recently had another ‘hickup’ in which they changed something leading to some data clients having hiccups. This includes GNOME applet Stocks Extension I had been running. There is a lively discussion on its issue #120 suggestions for example a curl wrapper (which then makes each access a new system call).
Separating data acquisition and presentation becomes an attractive alternative, especially given how the standard Python and R accessors to the Yahoo! Finance service continued to work (and how per post #36 I already run data acquisition). Moreoever, and somewhat independently, it occurred to me that the cute (and both funny in its pun, and very pretty in its display) ActivateLinux program might offer an easy-enough way to display updates on the desktop.
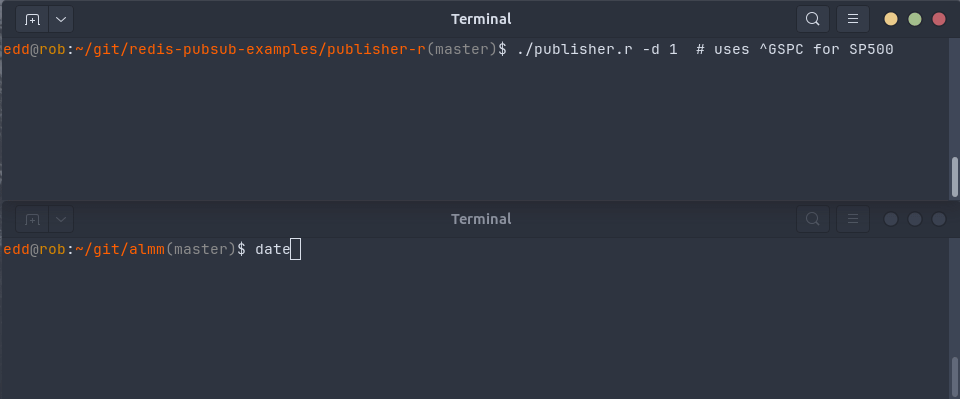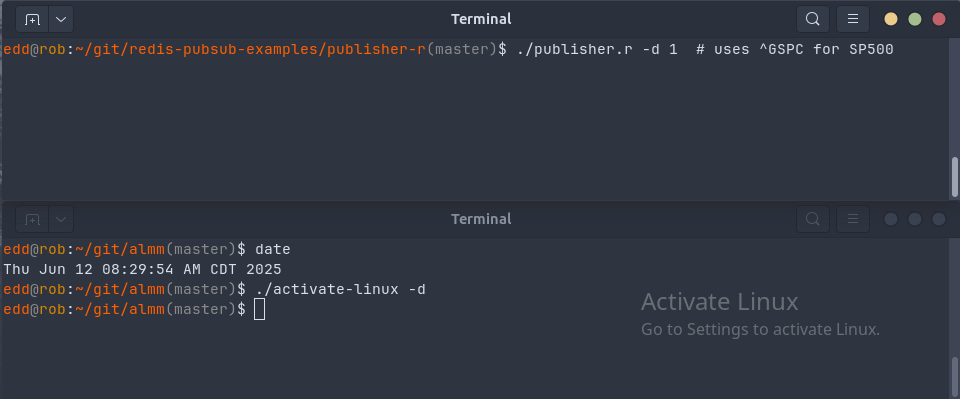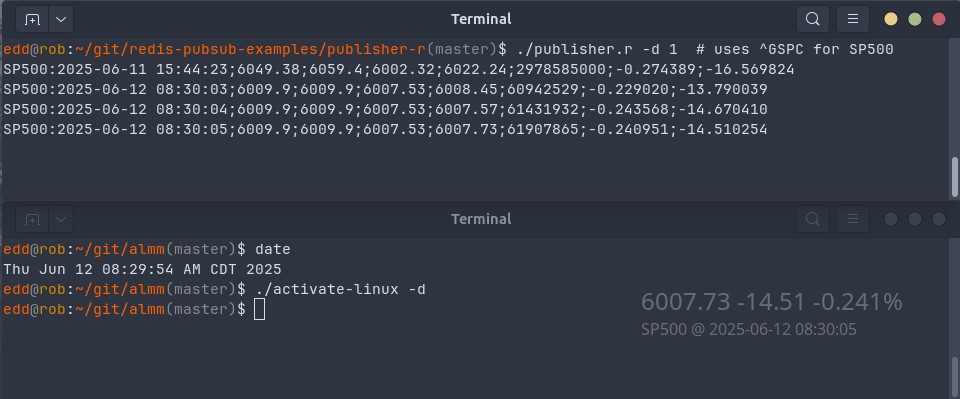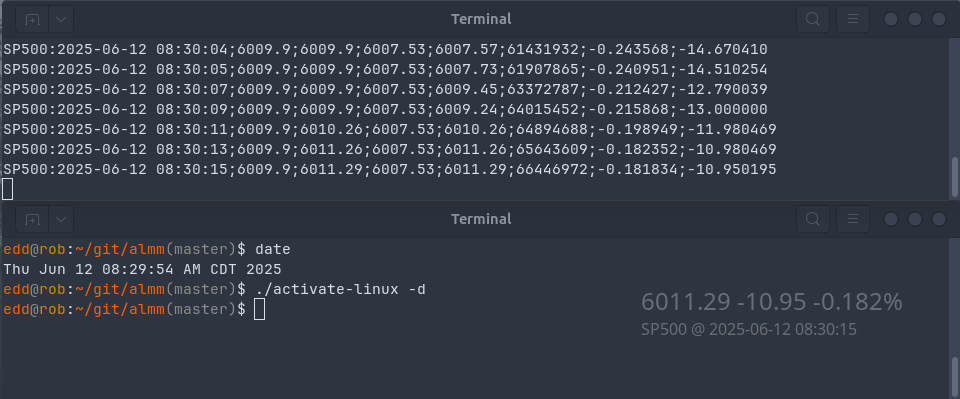
There were two aspects to address. First, the subscription side
needed to be covered in either plain C or C++. That, it turns out, is
very straightforward and there are existing documentation and prior
examples (e.g. at StackOverflow) as well as the ability to have an LLM
generate a quick stanza as I did with Claude. A modified variant is now
in the example
repo ‘redis-pubsub-examples’ in file subscriber.c.
It is deliberately minimal and the directory does not even have a
Makefile: just compile and link against both
libevent (for the event loop controlling this) and
libhiredis (for the Redis or Valkey connection). This
should work on any standard Linux (or macOS) machine with those two
(very standard) libraries installed.
The second aspect was trickier. While we can get Claude to modify the
program to also display under x11, it still uses a single controlling
event loop. It took a little bit of probing on my event to understand
how to modify (the x11 use of) ActivateLinux,
but as always it was reasonably straightforward in the end: instead of
one single while loop awaiting events we now first check
for pending events and deal with them if present but otherwise do not
idle and wait but continue … in another loop that also checks on the Redis or Valkey “pub/sub” events. So two thumbs up
to vibe coding
which clearly turned me into an x11-savvy programmer too…
The result is in a new (and currently fairly bare-bones) repo almm. It includes all
files needed to build the application, borrowed with love from ActivateLinux
(which is GPL-licensed, as is of course our minimal extension) and adds
the minimal modifications we made, namely linking with
libhiredis and some minimal changes to
x11/x11.c. (Supporting wayland as well is on the TODO list,
and I also need to release a new RcppRedis version
to CRAN as one currently needs
the GitHub version.)
We also made a simple mp4 video with a sound overlay which describes the components briefly:
Comments and questions welcome. I will probably add a little bit of
command-line support to the almm. Selecting the
symbol subscribed to is currently done in the most minimal way via
environment variable SYMBOL (NB: not SYM as
the video using the default value shows). I also worked out how to show
the display only one of my multiple monitors so I may add an explicit
screen id selector too. A little bit of discussion (including minimal Docker use around r2u) is also in issue
#121 where I first floated the idea of having StocksExtension
listen to Redis (or Valkey). Other suggestions are most
welcome, please use issue tickets at the almm repository.
This post by Dirk Eddelbuettel originated on his Thinking inside the box blog. If you like this or other open-source work I do, you can now sponsor me at GitHub.