Wicked Webapps with R, err, Wt
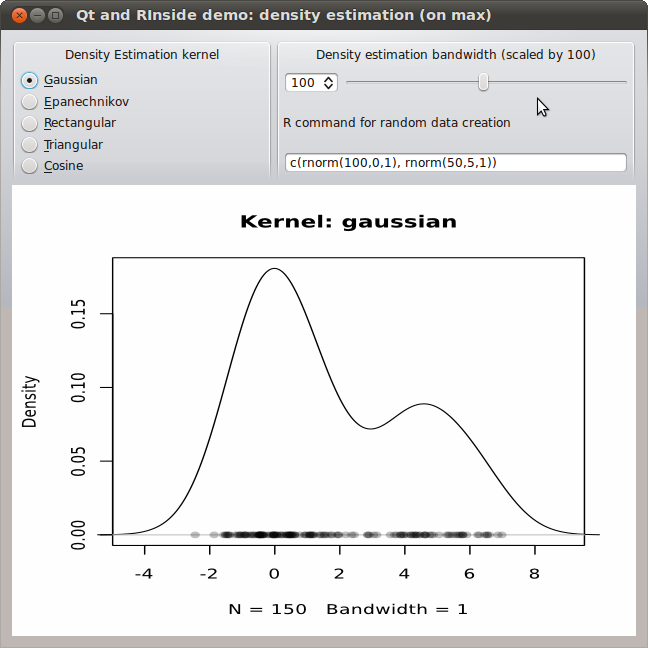
A few months ago, I had blogged about using R inside of Qt. This used our RInside package for embedding the statistical programming environment and language R inside of a C++ application, and further relies on our Rcpp package for R and C++ integration and object mapping.The example was simple yet powerful: a reimplementation of the standard GUI application of a standard density estimate. Here the user can pick a kernel density function from a selection, and also slide a bandwidth parameter. One nice addition was an entry field already populated with a simple expression for a mixture of Normals, allowing for arbitrary random distributions over which to estimate. The example is pretty (thanks to Qt), and was added to RInside with the last CRAN release 0.2.4. The blog post has a nice screenshot.
I had long wondered how to do something similar 'on the web'. Web integration and application frameworks are of course a dime a dozen: Just about any language offers this, with more or less ease. But I wanted something simple yet powerful and fast. And I did not like the idea of a multi-tier app, or of a multi-language mix. I remember having seen something about a web-application framework not unlike Qt, and studying the very useful Wikipedia web application framework comparison I re-discovered Wt (pronounced "Witty"). So there it is, using C++ which brings us ample performance, the ability to connect to a number of libraries and applications (which is important in my quantitatively-minded workd) and avoid the whole multi-tier, multi-language combination. The Wt website has a few more good reason why this may be a suitable idea; the toolkit also offers a very decent amount of features and is amply documented with a fair number of examples.
And after just a little bit poking around during two weekends, I now have the following webapp committed in the SVN repository of RInside, and it is all implemented in in less than two hundred (generously commented) lines of code.
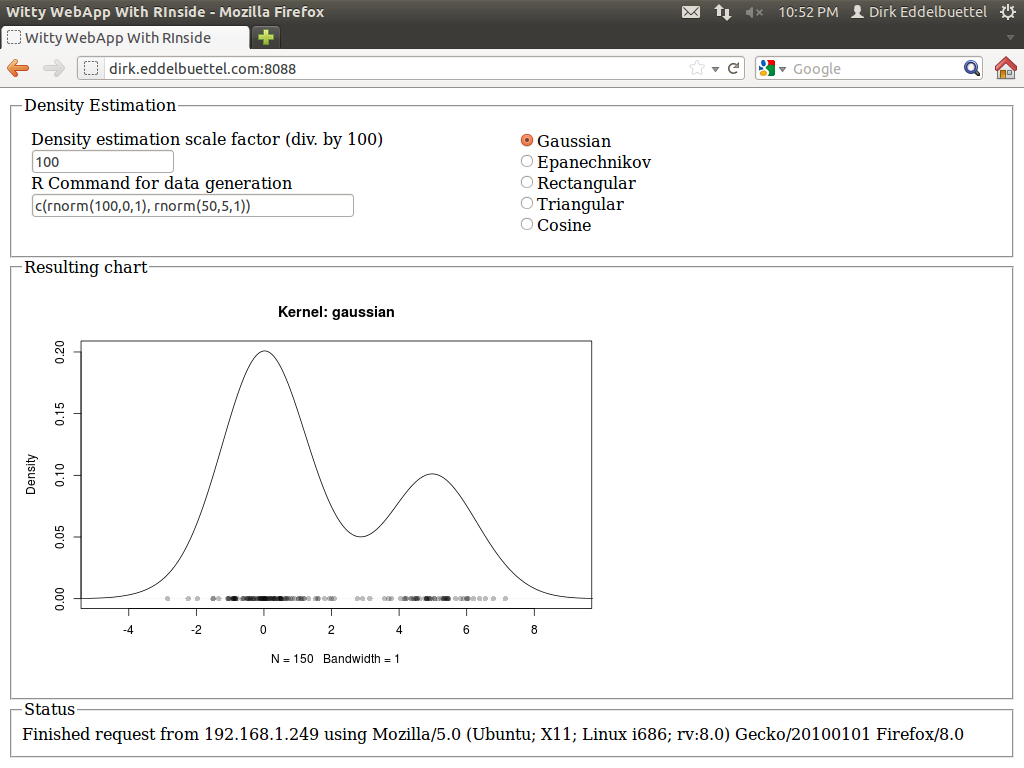
It is currently up and running at the address shown in the screenshot, so give it a go (though I may take it down at another point in time). I quite like it: The application is responsive: changes in the radio buttons (for the density), or the bandwidth, each trigger reestimation of the density, and a new and updated chart is displayed immediately with no noticeable delay---just like the desktop application did.
Best of all, the code logic is essentially unchanged from the Qt-based app. Signals and slots related events to actions, the layout is in terms of standard GUI boxen and containers. And best of all, I did not have to write a line of html, javascript, css or ajax: it is all handled by the Wt toolkit. I was able to drive the app from an Android phone, my tablet, various computers around the house, and had a few friends poke a stick at it from afar.
There is at one open issue. Wt launches new instances of the application object with each connection, which is a very clean model. That doesn't map perfectly with R (which is single-threaded) and RInside (which runs as a singleton for the same reason). So right now, each action sends its state back to the client. In other words, each clients own its parameters and well as vector of random numbers. Each new action sends these back to the app which passes it to R, launches the re-estimation and gets an updated chart back which is then shown by the the client. That is not perfect, and maybe a forking model as used by Simon's RServe would be better, though it would require a rewrite of RInside. Not sure if we get there anytime soon. And for simple applications not facing legions of concurrent users, the singleton should still work. It's a proof of concept at this point. Feedback welcome, and RInside and Rcpp questions should go to the rcpp-devel list as usual.